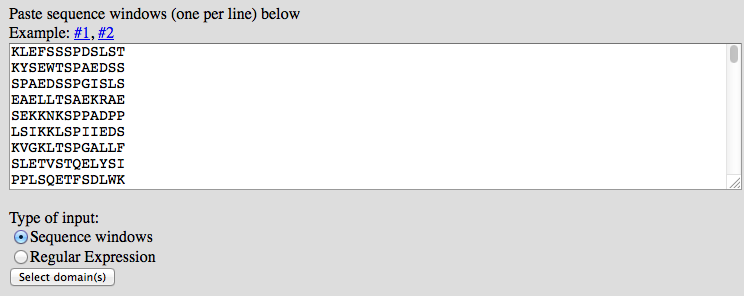
1) DoReMi accepts regular expressions (RegExp) or a set of peptides as input.
To use a RegExp, write it into the input field, select "Regular Expression" at the checkboxes and press submit.
To calculate a PSSM based on a set of know sites, paste the set of peptides into the input box, one per line. Select "Sequence windows" and press submit.
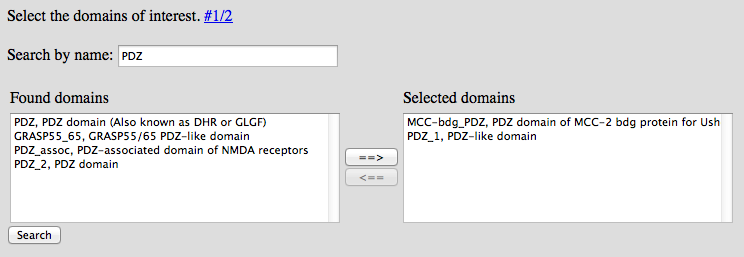
2) In the second step, interacting domains have to be selected.
To search for specific domains, write the (partial) name into the input box.
The left option box will show all available domains matching the search term. Select the domains of interest and press the "==>" button to move it to the right option box.
Domains will not be removed from the right box until manuall removal (selecting and pressing "<=="). Thereby, multiple domains can be added using different search terms.
When pressing submit, all domains in the right option list will be used to compute the association scores.
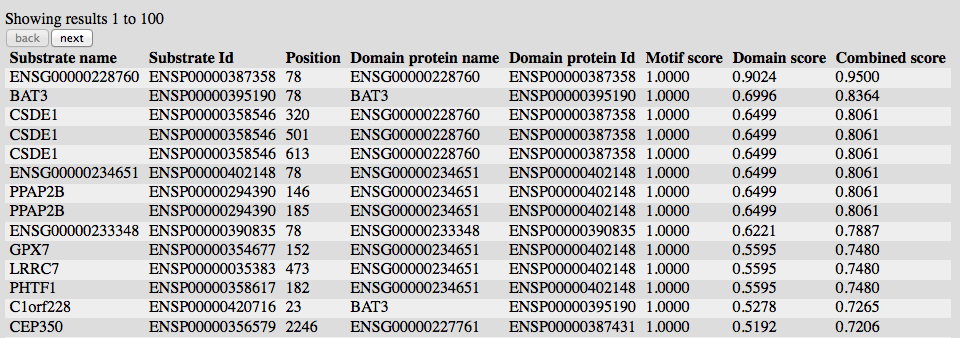
3) The results are presented in tabular form.
By default, the first 100 best scoring positions are shown, but using the two buttons on top, it is possible to go forth and back in steps of 100 lines.
Substrate and Position show he location of the found site. Domain protein lists the domain carrying the domain of interest which has been used to compute the association score.
In case a RegExp has been used to search for the initial site, the Motif score will be always 1.

 input motif data
input motif data
 select domains
select domains view results
view results